03:00
3 - What makes a model?
Machine learning with tidymodels
Your turn

How do you fit a linear model in R?
How many different ways can you think of?
lmfor linear modelglmnetfor regularized regressionkerasfor regression using TensorFlowstanfor Bayesian regressionsparkfor large data sets
Artwork by @allison_horst
To specify a model ![]()
- Choose a model
- Specify an engine
- Set the mode
To specify a model ![]()
To specify a model ![]()
- Choose a model
- Specify an engine
- Set the mode
To specify a model ![]()
To specify a model ![]()
To specify a model ![]()
- Choose a model
- Specify an engine
- Set the mode
To specify a model ![]()
To specify a model ![]()
All available models are listed at https://www.tidymodels.org/find/parsnip/
To specify a model ![]()
- Choose a model
- Specify an engine
- Set the mode
Your turn

Run the tree_spec chunk in your .qmd.
Edit this code so it creates a different model, such as linear regression.
05:00
All available models are listed at https://www.tidymodels.org/find/parsnip/
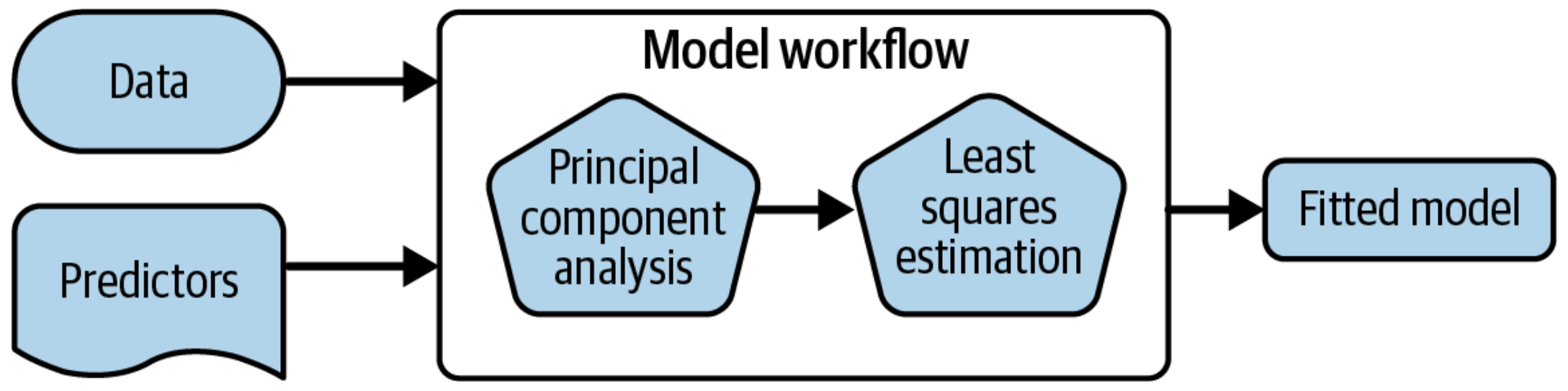
A model workflow
Workflows bind preprocessors and models
What is wrong with this?
Why a workflow()? ![]()
You can use other preprocessors besides formulas (more on feature engineering later!)
They can help organize your work when working with multiple models
Most importantly, a workflow captures the entire modeling process:
fit()andpredict()apply to the preprocessing steps in addition to the actual model fit
A model workflow ![]()
![]()
tree_spec <- decision_tree(mode = "regression")
tree_spec %>%
fit(rings ~ ., data = ring_train)
#> parsnip model object
#>
#> n= 3340
#>
#> node), split, n, deviance, yval
#> * denotes terminal node
#>
#> 1) root 3340 34681.9200 9.937425
#> 2) shell_weight< 0.16775 1146 5102.2900 7.584642
#> 4) shell_weight< 0.05325 242 484.3512 5.524793 *
#> 5) shell_weight>=0.05325 904 3316.2640 8.136062
#> 10) sex=infant 557 1432.8580 7.565530 *
#> 11) sex=female,male 347 1411.0660 9.051873 *
#> 3) shell_weight>=0.16775 2194 19922.2800 11.166360
#> 6) shell_weight< 0.35775 1588 11128.8300 10.587530
#> 12) shell_weight< 0.24925 679 3807.1960 9.948454
#> 24) shucked_weight>=0.24775 528 1773.1650 9.460227 *
#> 25) shucked_weight< 0.24775 151 1468.0930 11.655630 *
#> 13) shell_weight>=0.24925 909 6837.1710 11.064910
#> 26) shucked_weight>=0.39975 620 2638.9340 10.372580 *
#> 27) shucked_weight< 0.39975 289 3263.5220 12.550170 *
#> 7) shell_weight>=0.35775 606 6867.1680 12.683170
#> 14) shucked_weight>=0.55025 429 3609.9910 12.004660
#> 28) shell_weight< 0.579 382 2243.0990 11.607330 *
#> 29) shell_weight>=0.579 47 816.4255 15.234040 *
#> 15) shucked_weight< 0.55025 177 2580.9940 14.327680 *A model workflow ![]()
![]()
tree_spec <- decision_tree(mode = "regression")
workflow(rings ~ ., tree_spec) %>%
fit(data = ring_train)
#> ══ Workflow [trained] ════════════════════════════════════════════════
#> Preprocessor: Formula
#> Model: decision_tree()
#>
#> ── Preprocessor ──────────────────────────────────────────────────────
#> rings ~ .
#>
#> ── Model ─────────────────────────────────────────────────────────────
#> n= 3340
#>
#> node), split, n, deviance, yval
#> * denotes terminal node
#>
#> 1) root 3340 34681.9200 9.937425
#> 2) shell_weight< 0.16775 1146 5102.2900 7.584642
#> 4) shell_weight< 0.05325 242 484.3512 5.524793 *
#> 5) shell_weight>=0.05325 904 3316.2640 8.136062
#> 10) sex=infant 557 1432.8580 7.565530 *
#> 11) sex=female,male 347 1411.0660 9.051873 *
#> 3) shell_weight>=0.16775 2194 19922.2800 11.166360
#> 6) shell_weight< 0.35775 1588 11128.8300 10.587530
#> 12) shell_weight< 0.24925 679 3807.1960 9.948454
#> 24) shucked_weight>=0.24775 528 1773.1650 9.460227 *
#> 25) shucked_weight< 0.24775 151 1468.0930 11.655630 *
#> 13) shell_weight>=0.24925 909 6837.1710 11.064910
#> 26) shucked_weight>=0.39975 620 2638.9340 10.372580 *
#> 27) shucked_weight< 0.39975 289 3263.5220 12.550170 *
#> 7) shell_weight>=0.35775 606 6867.1680 12.683170
#> 14) shucked_weight>=0.55025 429 3609.9910 12.004660
#> 28) shell_weight< 0.579 382 2243.0990 11.607330 *
#> 29) shell_weight>=0.579 47 816.4255 15.234040 *
#> 15) shucked_weight< 0.55025 177 2580.9940 14.327680 *Your turn

Run the tree_wflow chunk in your .qmd.
Edit this code so it uses a linear model.
05:00
Predict with your model ![]()
![]()
How do you use your new tree_fit model?
Your turn

Run:
predict(tree_fit, new_data = ring_test)
What do you get?
03:00
Your turn

Run:
augment(tree_fit, new_data = ring_test)
What do you get?
03:00
The tidymodels prediction guarantee!
- The predictions will always be inside a tibble
- The column names and types are unsurprising and predictable
- The number of rows in
new_dataand the output are the same
Understand your model ![]()
![]()
How do you understand your new tree_fit model?
You can use your fitted workflow for model and/or prediction explanations:
Learn more at https://www.tmwr.org/explain.html
Understand your model ![]()
![]()
How do you understand your new tree_fit model?
Understand your model ![]()
![]()
How do you understand your new tree_fit model?
You can extract_*() several components of your fitted workflow: https://workflows.tidymodels.org/reference/extract-workflow.html
⚠️ Never predict() with any extracted components!
Deploy your model ![]()
How do you use your new tree_fit model in production?
Learn more at https://vetiver.rstudio.com
Deploy your model ![]()
How do you use your new model tree_fit in production?
library(plumber)
pr() %>%
vetiver_api(v)
#> # Plumber router with 2 endpoints, 4 filters, and 1 sub-router.
#> # Use `pr_run()` on this object to start the API.
#> ├──[queryString]
#> ├──[body]
#> ├──[cookieParser]
#> ├──[sharedSecret]
#> ├──/logo
#> │ │ # Plumber static router serving from directory: /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/library/vetiver
#> ├──/ping (GET)
#> └──/predict (POST)Learn more at https://vetiver.rstudio.com
Your turn

Run the vetiver chunk in your .qmd.
Check out the automated visual documentation.
05:00


